ORNi-PCR® to detect rare target sequences
Science & Technologies
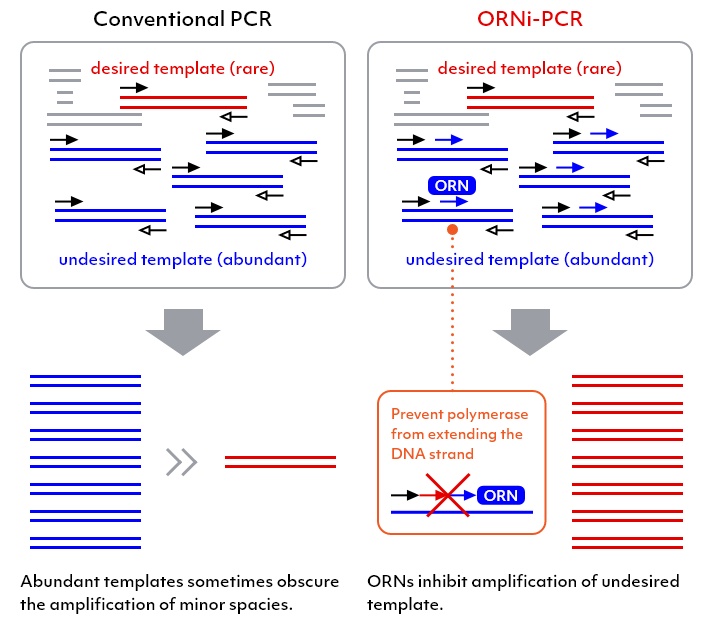
DNA sequence variants are known to be involved in the emergence of intractable diseases such as cancer. Various methods using PCR have been employed to detect the presence or absence of DNA mutations known to cause these diseases. However, conventional PCR-based methods have several limitations, such as sensitivity, cost, and ability to provide a definitive diagnosis. Oligoribonucleotide interference-PCR (ORNi-PCR®) has been invented to overcome these limitations.
The ORNi-PCR® technology is a method, in which an oligoribonucleotide (ORN, small RNA), an approximately 17-29 base RNA, to specifically inhibit the amplification of a DNA domain complementary and thus hybridized to the ORN in order to halt a PCR. Conversely, ORNi-PCR® enables the specific amplification of DNA with mutations in domains hybridized with an ORN. ORNi-PCR® does not always require information regarding mutation sequences if the target DNA sequence and mutation site to be suppressed for amplification are defined in advance. ORNi-PCR® has been reported to have the capacity to distinguish single base mutations with high precision and sensitivity and has thus been used for applications, such as detecting genome-edited cells, DNA methylation analysis, and bacterial flora analysis.
Original Reports
-
Fujita T, Nagata S, Fujii H: Oligoribonucleotide-mediated blockade of DNA extension by Taq DNA polymerases increases specificity and sensitivity for detecting single-nucleotide differences. Anal. Chem. (2023) 95, 3442-3451.
Link -
Fujita T, Nagata S, Fujii H: Protein or ribonucleoprotein-mediated blocking of recombinase polymerase amplification enables the discrimination of nucleotide and epigenetic differences between cell populations. Commun. Biol. (2021) 4, 988.
Link -
Fujita T, Nagata S, Yuno M, Fujii H: Sequence-specific inhibition of reverse transcription by recombinant CRISPR/dCas13a ribonucleoprotein complexes in vitro. Biol. Methods Protoc. (2021) 6, bpab009.
Link -
Shimizu T, Fujita T, Fukushi S, Horino Y, Fujii H: Discrimination of CpG methylation status and nucleotide differences in tissue specimen DNA by oligoribonucleotide interference-PCR. Int. J. Mol. Sci. (2020) 21, 5119.
Link -
Baba K, Fujita T, Tasaka S, Fujii H: Simultaneous detection of the T790M and L858R mutations in the EGFR gene by oligoribonucleotide interference-PCR. Int. J. Mol. Sci. (2019) 20, 4020.
Link -
Fujita T, Motooka D, Fujii H: Target enrichment from a DNA mixture by oligoribonucleotide interference-PCR (ORNi-PCR). Biol. Methods Protoc. (2019) 4, bpz009.
Link -
Fujita T, Yuno M, Kitaura F, Fujii H: A refined two-step oligoribonucleotide interference-PCR method for precise discrimination of nucleotide differences. Sci. Rep. (2018) 8, 17195.
Link -
Fujita T, Yuno M, Kitaura F, Fujii H: Detection of genome-edited cells by oligoribonucleotide interference-PCR. DNA Res. (2018) 25, 395-407.
Link -
Tanigawa N, Fujita T, and Fujii H: Oligoribonucleotide (ORN) interference-PCR (ORNi-PCR): a simple method for suppressing PCR amplification of specific DNA sequences using ORNs. PLoS One (2014) 9, e113345.
Link
Reviews
-
Shimizu T, Fujita T, and Fujii H: Oligoribonucleotide interference-PCR: principles and applications. Biol. Methods Protoc. (2022) 7 (1), bpac010.
Link


